Document Type
Article
Publication Date
3-1-2020
Abstract
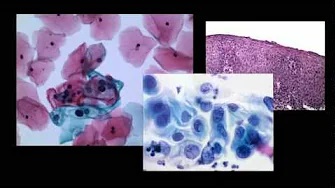
We have encountered pancreatic tumors with unique histologic features, which do not conform to any of the known tumors of the pancreas or other anatomical sites. We aimed to define their clinicopathologic features and whether they are characterized by recurrent molecular signatures. Eight cases were identified; studied histologically and by immunohistochemistry. Selected cases were also subjected to whole-exome sequencing (WES; n = 4), RNA-sequencing (n = 6), Archer FusionPlex assay (n = 5), methylation profiling using the Illumina MethylationEPIC (850k) array platform (n = 6), and TERT promoter sequencing (n = 5). Six neoplasms occurred in females. The mean age was 43 years (range: 26-75). Five occurred in the head/neck of the pancreas. All patients were treated surgically; none received neoadjuvant/adjuvant therapy. All patients are free of disease after 53 months of median follow-up (range: 8-94). The tumors were well-circumscribed, and the median size was 1.8 cm (range: 1.3-5.8). Microscopically, the unencapsulated tumors had a geographic pattern of epithelioid cell nests alternating with spindle cell fascicles. Some areas showed dense fibrosis, in which enmeshed tumor cells imparted a slit-like pattern. The predominant epithelioid cells had scant cytoplasm and round-oval nuclei with open chromatin. The spindle cells displayed irregular, hyperchromatic nuclei. Mitoses were rare. No lymph node metastases were identified. All tumors were positive for vimentin, CD99 and cytokeratin (patchy), while negative for markers of solid pseudopapillary neoplasm, neuroendocrine, acinar, myogenic/rhabdoid, vascular, melanocytic, or lymphoid differentiation, gastrointestinal stromal tumor as well as MUC4. Whole-exome sequencing revealed no recurrent somatic mutations or amplifications/homozygous deletions in any known oncogenes or tumor suppressor genes. RNA-sequencing and the Archer FusionPlex assay did not detect any recurrent likely pathogenic gene fusions. Single sample gene set enrichment analysis revealed that these tumors display a likely mesenchymal transcriptomic program. Unsupervised analysis (t-SNE) of their methylation profiles against a set of different mesenchymal neoplasms demonstrated a distinct methylation pattern. Here, we describe pancreatic neoplasms with unique morphologic/immunophenotypic features and a distinct methylation pattern, along with a lack of abnormalities in any of key genetic drivers, supporting that these neoplasms represent a novel entity with an indolent clinical course. Given their mesenchymal transcriptomic features, we propose the designation of "sclerosing epithelioid mesenchymal neoplasm" of the pancreas.
Recommended Citation
Basturk O, Weigelt B, Adsay V, Benhamida JK, Askan G, Wang L, Arcila ME, Zamboni G, Fukushima N, Gularte-Mérida R, Da Cruz Paula A, Selenica P, Kumar R, Pareja F, Maher CA, Scholes J, Oda Y, Santini D, Doyle LA, Petersen I, Flucke U, Koelsche C, Reynolds SJ, Yavas A, Deimling AV, Reis-Filho JS, Klimstra DS. Sclerosing epithelioid mesenchymal neoplasm of the pancreas - a proposed new entity. Mod Pathol. 2020 Mar;33(3):456-467. doi: 10.1038/s41379-019-0334-5. Epub 2019 Aug 5. PMID: 31383964; PMCID: PMC7000300.